|
NeuroNames Statistics
(as of June 1, 2004)
The NeuroNames nomenclature contains about 15000 terms, 2000 of which are acronyms. It
allows a complete description of the primate brain in terms of about 900
structures (approximately 580 primary volumetric structures, 130 superstructures
and 180 superficial structures). Using NeuroNames, any point in the brain
can be located unambiguously to a single primary structure. Thus, mastery
of the names, definitions and relationships among
about 900 structures should provide one with a working knowledge of the
basic neuroanatomy of the brain. Since each of the structures in
the Hierarchy has a default English name and a default Latin name, that
portion of the nomenclature accounts for 1720 of the total terms.
The remaining terms represent ancillary structures or are synonyms in seven languages.
With 1500 ancillary structures, synonyms account for almost two-thirds of
the total neuroanatomical vocabulary.
Structures (total 2402)
-
Hierarchy Structures - 891
-
Primary Volumetric Structures - 580
-
Superstructures - 127
-
Superficial Structures - 184
-
Ancillary Structures - 1511
Terms (total 15373)
-
Default English and Latin Terms in the Hierarchy - 1782
-
Default Ancillary Terms - 1511
-
Synonyms - 9985
-
Acronyms - 2095
BrainInfo Usage
-
240 visitors/day
-
9,000 pages viewed/day
Template Atlas
To explore the Template Atlas Open BrainInfo Again in a separate window and navigate according to the instructions below.
The Template Atlas of BrainInfo is a set of drawings of the brain that includes four cortical views and 58 coronal sections of the brain of the longtailed macaque (Macaca fascicularis). Based on three animals, it is an updated version of the atlas that we first published in 1996. For a detailed description of the methods used in its production see Martin & Bowden, 1996 and Section 3.2: Methods in Martin & Bowden, 2000.
The digital format of the atlas in BrainInfo has been adapted to allow maximal flexibility in display of the templates, e.g., with or without the structure names, the shading of gray matter, the estimated structural boundaries, the stereotaxic scales, etc. Templates in a format suitable for downloading to image processing software to map data for publication are available on a compact disk (CD-ROM) that accompanies the hard copy version of the atlas: Primate Brain Maps: Structure of the Macaque Brain (Martin & Bowden, 2000).
To locate a structure by browsing the Template Atlas of BrainInfo, click 'Search Atlas' button. The images below will appear.
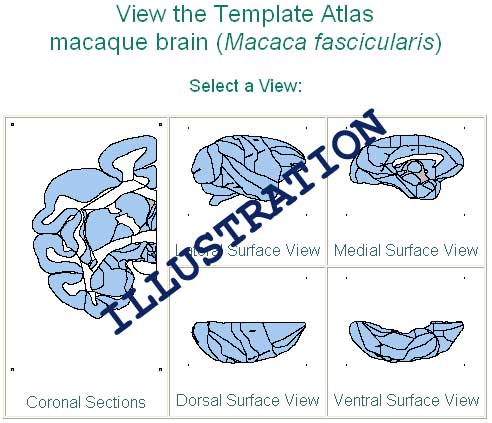
If the structure you want is part of the cortex, click the appropriate cortical view. If it is a deep structure, click the cross-sectional image. For cross-sectional views the Template Atlas will show you about 30 small images of the brain in cross-section ranging from the back to the front of the brain. Click the image that appears to be closest to the section that interests you and then use the
'Go Anterior' button and 'Go Posterior' button to reach the exact section you want.
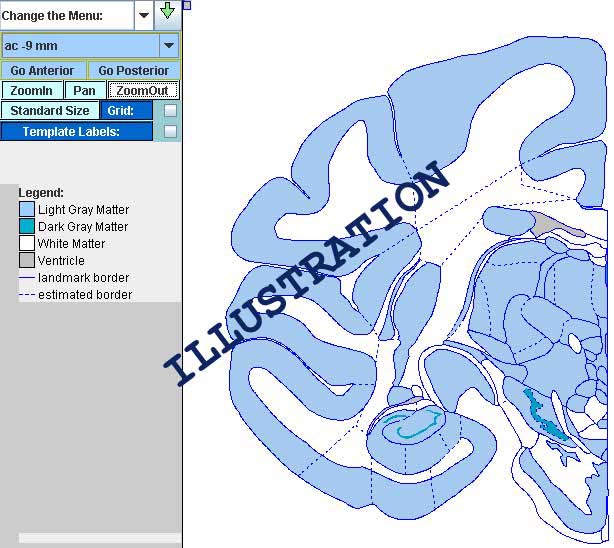
To zoom on the whole section, click the 'Zoom In' button and then click anywhere on the atlas. If you want to zoom on a particular part of the image, click
'Zoom In' and then click and drag to create a rectangle that contains the area of interest.
To zoom out on the whole section, click the 'Zoom Out' button and then click anywhere on the atlas. If you want to zoom out and still view only a part of the images, click
'Zoom Out' and then click and drag to create a rectangle that contains the area of interest.
To label structures click on  . .
Stereotaxis
As in human neurosurgery, the accuracy of brain surgery in nonhuman primates can be greatly aided by noninvasive imaging with magnetic resonance (MR) and ventriculography. These techniques allow the surgeon to direct instruments to their targets based on internal landmarks, such as the anterior and posterior commissures, as well as external landmarks, such as the ear canals, orbits of the eyes, top of the skull, etc. The Template Atlas of BrainInfo was developed and scaled particularly for use with such imaging techniques, as well as for conventional
stereotaxis.
MRI Stereotaxis: Again, as in human stereotaxis, the primary coordinate system of the Template Atlas is based on internal landmarks. The base plane is perpendicular to the midline and passes through the bicommissural line, i.e., the line defined by the centers of the anterior commissure and the posterior commissure. Zero of all three dimensions is located at the center of the anterior commissure.
Users able to take advantage of imaging techniques, such as magnetic resonance imaging (MRI) or ventriculography to identify the commissures can reduce stereotaxic error by up to 80% or more by using these internal landmarks (Dubach et al., 1985). Accuracy can be increased further by warping the appropriate atlas template to the coronal MRI plane of the target site and using local landmarks that exist in both the atlas and the MRI to estimate the coordinates of the region of interest. (See Section 4.2: Mapping templates to individuals for stereotaxis in Martin & Bowden, 2000.)
Conventional Stereotaxis: Users with no access to noninvasive techniques can obtain the same degree of accuracy with the Template Atlas that is obtained with the conventional atlases of the longtailed macaque brain, which are based on external, cranial landmarks (Szabo & Cowan,1984; Shantha et al.,1968; see Applicability of the template atlas to other primate species, Chapter 5 in Martin & Bowden, 2000). The interaural line, i.e., the line connecting the tips of the earbars of the stereotaxic instrument, and the top of the brain are the most reliable landmarks for use during brain surgery without imaging. To obtain the estimated location in the experimental brain of any point in the Template Atlas using these external landmarks: 1) estimate the AP (anterior-posterior) coordinate by adding 17mm to the AP coordinate of the Template Atlas, e.g., a point in section ac -15 of the atlas is located at eb +2, i.e., 2 mm anterior to the earbars (eb); 2) determine the distance of the point from the top of the brain in the experimental animal by subtracting the point's DV (dorsal-ventral) coordinate in the atlas from 19 mm, i.e., from the average height of the top of the atlas brain above the base plane; 3) the ML (medial-lateral) coordinate in the experimental brain is the same as in the atlas, since both are measured from the midline.
Stereotaxis by Ventriculography: In addition to correcting for the height of the brain above the orbitomeatal plane as indicated above, with a lateral ventriculogram one can compensate for variation in the orientation of the brain. This is done by adjusting the eyebars up or down to make the orbitomeatal plane of the instrument parallel the bicommissural line, and to register the stereotaxic space to that internal referent instead to the top of the brain. In this case one targets atlas structures in the animal by adding the atlas AP and DV coordinates to the corresponding distances of the anterior commissure from earbar zero.
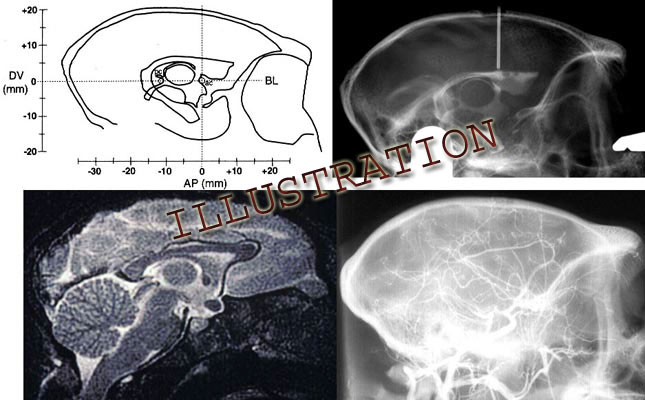
A graphical method for calculating the distance that the height of the eyebars should be adjusted is presented in Dubach, 1991. Equations for making the adjustment from MRI landmarks are presented in Martin and Bowden, 1996. Making this adjustment can greatly reduce the error of targeting based on cranial landmarks alone depending on the angle by which the orbitomeatal plane deviates from that of the bicommissural line and the distance of the target from the center of the anterior commissure (Dubach et al., 1985).
The graphical solution in the publication cited contains a misprint in the caption of Fig.2. A5 should equal the square root of (A32 - HA2), not the square root of (A32 x HA2).
NeuroMaps
Work in progress.
History of BrainInfo
BrainInfo grew out of a concept born with the appearance of the first desktop computers in the mid-1970s. For an informal history see CENTER UPDATE: Issue 2, Spring 2001.
| 1991 |
Version 1 of NeuroNames programmed in HyperCard, copyrighted and issued
on diskette for the Macintosh. |
| 1993 |
Version 1 of the NeuroNames English Hierarchy incorporated as a source vocabulary
of the Metathesaurus of the Unified Medical Language System (UMLS) of The
National Library of Medicine (Source Vocabulary UWA92) |
| 1995 |
NeuroNames Hierarchy peer reviewed and modified for publication (Bowden,
1995). Superstructures pruned to eliminate disputed categories; primary
structures added and subtracted to conform to 12 recent texts and atlases. |
| 1996 |
Version 2 of NeuroNames English Hierarchy incorporated into the Metathesaurus
of UMLS (Source Vocabulary NEU95). |
| 1996 |
Template Atlas of the Primate Brain with structures labeled according
to NeuroNames peer-reviewed and published in condensed journal format (Martin,
1996). |
| 1997 |
Template Atlas website established with downloadable templates for
plotting neuroanatomical data. |
| 1997 |
First edition of the complete Template Atlas published by Primate Information Center (Martin,
1997). |
| 1998 |
Version 3 of the NeuroNames Hierarchy. Major changes were to clarify
structures found only in the human or only in the nonhuman primate brain,
to include subdivisions of some structures that were previously primary
structures, and to simplify some of the more cumbersome standard terms,
e.g., "amygdala" for "amygdaloid nuclear complex." |
| 1998 |
Version 3 of NeuroNames English Hierarchy submitted for incorporation
into 1999 Metathesaurus of UMLS (NEU99). |
| 1998 |
Semantic network of NeuroNames extended to define several hundred brain
structures in terms of the standard terminology of the Hierarchy and to
provide synonyms for structures in the rat brain regarded as morphologically
homologous to structures in the Hierarchy (Bowden,
1997). |
| 1998 |
NeuroNames website established with Web interface to the Digital
Anatomist to show Hierarchy structures in the human brain and with internal interface
to the Template Atlas of the Primate Brain to show structures in the macaque
brain. |
| 2000 |
NeuroNames Hierarchy updated and about 4000 synonyms for hierarchy terms incorporated into the UMLS (NEU99).
|
|
2000 | NeuroNames and Template Atlas, including templates for downloading, incorporated into Primate Brain Maps: Structure of the Macaque Brain, a book and compact disk combination issued by Elsevier Science.
|
| 2001 | NeuroNames and Template Atlas website incorporated into BrainInfo website; symbolic information transferred to MS Access database; image information transferred to MapObjects (ESRI/ArcInfo) and Java software; server programmed in Visual Basic; links established to PubMed (NLM), to more than 300 brain images at other websites: Digital Anatomist (University of Washington); LONI (UCLA); Whole Brain Atlas (Harvard), to a site that describes the cells in specific structures (SenseLab/Yale) and to sites illustrating the species studied (Wisconsin RPRC, Primate Gallery, Washington RPRC). Other additions: the ability to search in Italian (1000 terms), Spanish (660 terms), and Russian (420 terms); a stereotaxic brainstem atlas of the rhesus macaque in sitting posture; PubMed citation counts for all English and Latin synonyms. Average use: 34 visitors/day viewing 1100 pages/day.
|
| 2002 | First NeuroMaps incorporated: capability for display of overlays on the Template Atlas to compare macaque cortical areas as defined by Brodmann (1909), Mauss (1908), the Vogts (1919), and Walker (1940); NeuroNames-2002 CD distributed to 15 other websites for use as neuroanatomical index or linking into BrainInfo; links established to Mouse Brain Library (University of Tennessee) to illustrate 300 structures in the mouse brain; DICTIONARY added to the sidebar, so when users are sent to another website they can look up unfamiliar terms without leaving that website; Russian structure names increased to 930, Spanish to 790, Indonesian to 1139. Average use: 46 visitors/day viewing 1300 pages/day.
|
| 2003 | Brodmann's areas in human linked to illustrations of Areas of Cortex Involved in Language (University of Washington) and to 3-D reconstruction of human cortex LONI (UCLA); COMMENTS capability added to allow visitors to read and submit comments linked to every fact recorded in BrainInfo; German structure names incorporated (711 terms); NeuroNames reprogrammed in XML to allow generation of Hierarchy on the fly. Survey of first 10,000 visitors to BrainInfo showed reasons for visiting were research interest: 35%, educational: 35%, curiosity: 20%, clinical interest: 10%. Average use: 120 visitors/day viewing 4300 pages/day.
|
| 2004 |
Addition of Percent Overlap tool that allows one to select a cortical area, obtain a list of structures that overlap it and view their percents overlap; addition of NeuroMaps of 38 cytoarchitectonic areas of macaque cortex by von Bonin and Bailey (1947) and photomicrographs of the internal structure of 150 cortical areas; interoperability established with the Brain Architecture Management System (BAMS, USC, Los Angeles) to display connectivity among brain structures based on some 10,000 reports from studies in the rat; links established to more than 700 high resolution photomicrographs of the macaque brain in three planes of section at Brain Atlas Project (Center for Neuroscience, UC Davis), 50 illustrations of cortical areas in flat maps, inflated maps and surface views at Van Essen Lab (Washington University, St. Louis) and 120 illustrations of human brainstem and spinal cord structures at Medical Neuroscience website (Loyola University, Chicago). Addition of some 1700 neuroanatomical terms and acronyms from the rat brain atlas of Swanson-1998 and the mouse brain atlases of Hof-2000 and Paxinos-2001. Average use: 240 visitors/day viewing 9000 pages/day.
|
| 2005 |
Links established to: Van Essen Lab (Washington University, St. Louis) for connectivity information on 50 structures; images of 59 structures in Atlas of the Human Brain (Heinrich Heine University, Duesseldorf, Germany); gene expression in 122 structures at GENSAT (gene expression database, Rockefeller University and NLM); 383 photomicrographs of structures in macaque brain slide collections at BrainMaps (University of California, Davis); 30 photomicrographs of brainstem structures at the Washington National Primate Research Center (Seattle); 60 illustrations of brain structures at YUMC (Yonsei University, Seoul, Korea); 35 illustrations of cerebellar structures at Atlas of Human Anatomy (Sinel’nikov, Russia); 235 nissl-stained sections of human brain at the Brain Biodiversity Bank (Michigan State University). A new section – Tools & Methods - was added to BrainInfo. It includes nomenclatures of canonical atlases and NeuroNames for human and macaque neuroanatomy, Primate NeuroMaps (individual templates of macaque brain that can be downloaded for publication or for warping), and methods used to create a 3-D brain atlas. Average use: 230 visitors/day viewing 15,000 pages/day. |
| 2006 |
Updated the 2D Macaque Brain Atlas to highlight structures and follow them through all applicable sections, and to display overlap between structures. Links established for display of 286 mouse brain structures in coronal sections, 303 in sagittal sections at the Allen Brain Atlas (Seattle); 87 dissected structures at the Center for Biostructure, Medical University, Warsaw; 40 structures at the Institute of Anatomy, University of Vienna; 35 structures of the fetal mouse brain at GenePaint (Max-Planck Institute, Hannover, Germany); 32 structures at the Anatomical Atlas where is this?; and 65 illustrations at other websites. Completed definitions, photomicrographs and mapping to the Macaque Brain Atlas of cortical areas by Brodmann-1909 (30 areas), Mauss-1908 (31), Vogts-1919 (29), Walker-1940 (13), Bonin-1947 (35). Tools and Methods section expanded to include Nomenclature for the rat brain from the Swanson-2004 atlas. The website servers were updated to improve atlas speed, reliability and security, and BrainInfo acquired a shorter new address: www.braininfo.org. Average use: 330 visitors/day viewing 17,700 pages/day. |
| 2007 |
Definitions added for anatomical planes and some 400 neuroanatomical terms, including 300 terms in French. Links established for illustration of structures at: Nature Neuroscience; Keio University; University of Portsmouth; Human Psychophysiology; Brain: a Journal of Neurology. Linked to Gray's Anatomy on Wikipedia for the illustrations of Brodmann areas and to Histologischer Atlas im Internet for the illustrations of spinal cord structures. Established interconnectivity with Mouse Genome Informatics (MGI) for information on gene expression (92 structures). Expanded Tools & Methods section by adding “NeuroNames 2004: Mouse and Rat Brain Atlas Nomenclatures.” Average use: 384 visitors/day viewing 24,604 pages/day. |
|